The DISOclust Server Results
Please cite the following paper:McGuffin, L. J. (2008) Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics, 24, 586-587. PubMed
| DISOclust results for vjn7v89dg6iuk1jf | Help | |||

| |||
| Click here to download the plot above in PostScript. | |||
| The DISOclust method is based on the analysis of the variation in nFOLD3 models using ModFOLDclust. The nFOLD3 models and ModFOLDclust results are shown below. | |||
| ModFOLDclust results for vjn7v89dg6iuk1jf | Help | |||
| Model name (click for link to template PDB summary) | Model quality score | Residue error plot (click image for large version) | 3D view of residue error (click image for large version) |
| 2kpoa_spk2_TS4 | 0.1105 |
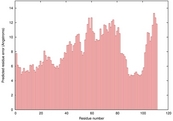 |
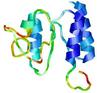 |
| 2kpoa_sp3_TS5 | 0.1105 |
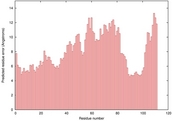 |
 |
| 2k4na_spk2_TS1 | 0.1061 |
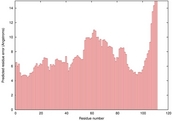 |
 |
| 2k4na_sp3_TS1 | 0.1061 |
 |
 |
| 2k4nA_HHsearch_TS1 | 0.1046 |
 |
 |
| multi_HHsearch_TS1 | 0.1031 |
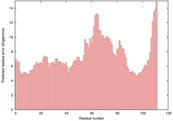 |
 |
| 1p3wb_spk2_TS9 | 0.1023 |
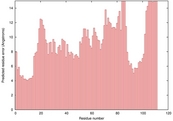 |
 |
| 2l69a_sp3_TS3 | 0.0992 |
 |
 |
| 2kfpa_sp3_TS9 | 0.0910 |
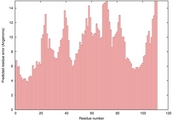 |
 |
| 1q7la_spk2_TS8 | 0.0781 |
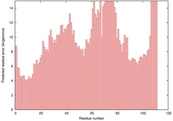 |
 |
| 2kj8a_sp3_TS6 | 0.0728 |
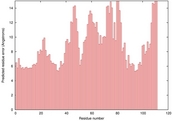 |
 |
| 2kiwa_sp3_TS10 | 0.0652 |
 |
 |
| 1fovA_HHsearch_TS2 | 0.0504 |
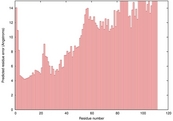 |
 |
| 1nupA_HHsearch_TS9 | 0.0453 |
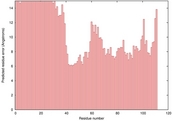 |
 |
| 3ic4A_HHsearch_TS5 | 0.0432 |
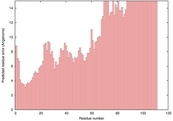 |
 |
| 2kjga_spk2_TS10 | 0.0432 |
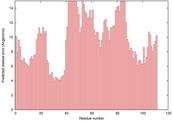 |
 |
| 1s2oa_spk2_TS3 | 0.0410 |
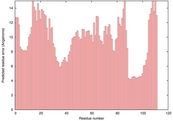 |
 |
| 2ky4a_spk2_TS6 | 0.0405 |
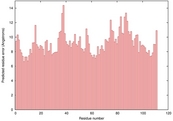 |
 |
| 1yozA_HHsearch_TS6 | 0.0389 |
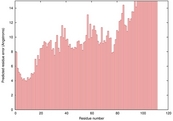 |
 |
| 2wciA_HHsearch_TS7 | 0.0361 |
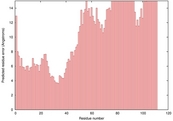 |
 |
| 1oiha_spk2_TS2 | 0.0349 |
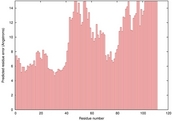 |
 |
| 2kena_spk2_TS5 | 0.0289 |
 |
 |
| 2kena_sp3_TS2 | 0.0289 |
 |
 |
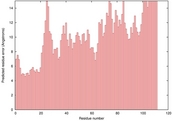
| 1r7hA_HHsearch_TS3 | 0.0266 |
 |
 |
| 2xggA_HHsearch_TS4 | 0.0238 |
 |
 |
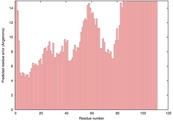
| 2ht9A_HHsearch_TS10 | 0.0206 |
 |
 |
| 1g51a_spk2_TS7 | 0.0131 |
 |
 |
| 2k5qa_sp3_TS4 | 0.0075 |
 |
 |
| 2kqvA_HHsearch_TS8 | 0.0073 |
 |
 |
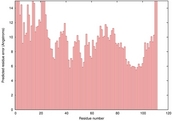
| 2hjqa_sp3_TS7 | 0.0068 |
 |
 |
| 2jxta_sp3_TS8 | 0.0000 |
 |
 |