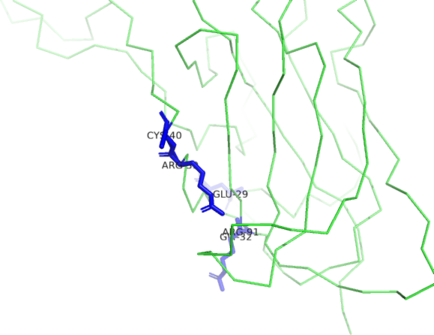
PyMOL generated image of ligand binding residues prediction for H1135_A
Click here to download PDB file of this model with the superposition of all identified ligands.
Predicted ligand binding residues are shown as blue sticks in the image above.
Binding site: 39, 40; 29, 32, 91
Most likely ligands at each site (Type): ASN; TYR
Centroid ligands at each site (TypeID): ASN206; PRO223
All ligands in clusters (Type-Frequency): TYR-1, ALA-1, CYS-1, ASN-2, HIS-1, PRO-1, PHE-1, SER-1; TYR-1, LEU-1, ASN-1, PRO-1, ARG-1
Likely+centroid ligands at each site: ASN206; TYR218
EC numbers:
GO terms:
JSmol view of ligand binding residues prediction for H1135_A
The server makes use of the FunFOLDQA Quality Assessment protocol for improved prediction selection:
Roche, D. B., Buenavista, M. T. & McGuffin, L. J. (2012) FunFOLDQA: A Quality Assessment Tool for Protein-Ligand Binding Site Residue Predictions. PLoS ONE, 7, e38219. PubMed
BDTalign = 0.06112164553682627
Identity = 0.0
Rescaled BLOSUM62 = 0.2
Equivalent Residue Ligand Distance = 0.044757303
Model Quality = 0.7598174
Predicted BDT score = 0.33610585
Predicted MCC score = 0.16636346
r=ARG; n=39; a=NH1; | I=0.00000; O=0.00000; N=0.00000; P=0.22902; | LEU=0.03163; TYR=0.00034; ALA=0.15947; CYS=0.01788; ASN=0.00265; SER=0.01704;
r=ARG; n=39; a=C; | I=0.00000; O=0.00000; N=0.00000; P=0.30469; | LEU=0.00388; ALA=0.01045; CYS=0.03958; ASN=0.14863; PHE=0.00894; SER=0.09320;
r=ARG; n=39; a=O; | I=0.00000; O=0.00000; N=0.00000; P=0.33529; | LEU=0.00311; ALA=0.00611; CYS=0.01791; ASN=0.19956; PHE=0.01910; ARG=0.00264; SER=0.08686;
r=ARG; n=39; a=CB; | I=0.00000; O=0.00000; N=0.00000; P=0.16379; | LEU=0.00339; ALA=0.03375; CYS=0.05122; ASN=0.03095; PHE=0.00048; SER=0.04400;
r=ARG; n=39; a=CA; | I=0.00000; O=0.00000; N=0.00000; P=0.22831; | LEU=0.00413; ALA=0.01780; CYS=0.02943; ASN=0.10319; PHE=0.00627; SER=0.06748;
r=ARG; n=39; a=CD; | I=0.00000; O=0.00000; N=0.00000; P=0.19062; | LEU=0.01050; ALA=0.09224; CYS=0.03539; ASN=0.01757; SER=0.03493;
r=ARG; n=39; a=CG; | I=0.00000; O=0.00000; N=0.00000; P=0.11322; | LEU=0.00232; ALA=0.04100; CYS=0.03135; ASN=0.01603; SER=0.02252;
r=ARG; n=39; a=N; | I=0.00000; O=0.00000; N=0.00000; P=0.17808; | LEU=0.00051; ALA=0.00472; CYS=0.00695; ASN=0.11351; PHE=0.02102; SER=0.03135;
r=ARG; n=39; a=NE; | I=0.00000; O=0.00000; N=0.00000; P=0.14409; | LEU=0.00543; ALA=0.10392; CYS=0.01724; ASN=0.00467; SER=0.01282;
r=ARG; n=39; a=CZ; | I=0.00000; O=0.00000; N=0.00000; P=0.20013; | LEU=0.01005; ALA=0.16540; CYS=0.01328; ASN=0.00163; SER=0.00977;
r=ARG; n=39; a=NH2; | I=0.00000; O=0.00000; N=0.00000; P=0.16198; | LEU=0.00232; ALA=0.15389; CYS=0.00385; SER=0.00191;
r=CYS; n=40; a=SG; | I=0.00000; O=0.00000; N=0.00000; P=0.36460; | LEU=0.02306; TYR=0.00209; ALA=0.04012; CYS=0.12346; ASN=0.02294; SER=0.15294;
r=CYS; n=40; a=CB; | I=0.00000; O=0.00000; N=0.00000; P=0.28229; | LEU=0.00267; ALA=0.01131; CYS=0.12392; ASN=0.04959; SER=0.09480;
r=CYS; n=40; a=N; | I=0.00000; O=0.00000; N=0.00000; P=0.27409; | LEU=0.00271; ALA=0.01264; CYS=0.07536; ASN=0.09348; PHE=0.00210; SER=0.08779;
r=CYS; n=40; a=CA; | I=0.00000; O=0.00000; N=0.00000; P=0.28570; | LEU=0.00120; ALA=0.00495; CYS=0.07414; ASN=0.11677; PHE=0.00144; ARG=0.00078; SER=0.08642;
r=CYS; n=40; a=C; | I=0.00000; O=0.00000; N=0.00000; P=0.21705; | ALA=0.00031; CYS=0.03785; ASN=0.13868; PHE=0.00203; ARG=0.00067; SER=0.03752;
r=CYS; n=40; a=O; | I=0.00000; O=0.00000; N=0.00000; P=0.19196; | CYS=0.02435; ASN=0.14125; PHE=0.00100; ARG=0.00215; SER=0.02322;
r=GLU; n=29; a=OE1; | I=0.00000; O=0.00000; N=0.00000; P=0.26982; | TYR=0.02651; LEU=0.23615; PRO=0.00189; PHE=0.00334; ARG=0.00193;
r=GLU; n=29; a=CD; | I=0.00000; O=0.00000; N=0.00000; P=0.25476; | TYR=0.04723; LEU=0.20639; PRO=0.00037; PHE=0.00040; ARG=0.00037;
r=GLU; n=29; a=CG; | I=0.00000; O=0.00000; N=0.00000; P=0.27795; | TYR=0.10808; LEU=0.16834; PRO=0.00073; ARG=0.00080;
r=GLU; n=29; a=OE2; | I=0.00000; O=0.00000; N=0.00000; P=0.19410; | TYR=0.02460; LEU=0.16950;
r=GLU; n=29; a=CA; | I=0.00000; O=0.00000; N=0.00000; P=0.16282; | TYR=0.12551; LEU=0.03264; PRO=0.00467;
r=GLU; n=29; a=O; | I=0.00000; O=0.00000; N=0.00000; P=0.27105; | TYR=0.17063; LEU=0.04538; PRO=0.05504;
r=GLU; n=29; a=CB; | I=0.00000; O=0.00000; N=0.00000; P=0.17669; | TYR=0.09828; LEU=0.07774; PRO=0.00067;
r=GLU; n=29; a=C; | I=0.00000; O=0.00000; N=0.00000; P=0.20112; | TYR=0.14498; LEU=0.03292; PRO=0.02322;
r=GLU; n=29; a=N; | I=0.00000; O=0.00000; N=0.00000; P=0.09644; | TYR=0.08705; LEU=0.00800; PRO=0.00138;
r=GLY; n=32; a=O; | I=0.00000; O=0.00000; N=0.00000; P=0.25760; | TYR=0.18634; LEU=0.00324; ASN=0.00101; PRO=0.06055; ARG=0.00645;
r=GLY; n=32; a=C; | I=0.00000; O=0.00000; N=0.00000; P=0.27322; | TYR=0.18449; LEU=0.00476; PRO=0.08261; ARG=0.00136;
r=GLY; n=32; a=CA; | I=0.00000; O=0.00000; N=0.00000; P=0.35512; | TYR=0.17977; LEU=0.01164; PRO=0.16329; ARG=0.00041;
r=GLY; n=32; a=N; | I=0.00000; O=0.00000; N=0.00000; P=0.29098; | TYR=0.12735; LEU=0.02491; PRO=0.13872;
r=ARG; n=91; a=CB; | I=0.00000; O=0.00000; N=0.00000; P=0.10511; | TYR=0.02224; LEU=0.00049; ASN=0.00049; ARG=0.08189;
r=ARG; n=91; a=CD; | I=0.00000; O=0.00000; N=0.00000; P=0.29910; | TYR=0.07038; LEU=0.00029; ASN=0.02480; ARG=0.20364;
r=ARG; n=91; a=NH1; | I=0.00000; O=0.00000; N=0.00000; P=0.44542; | TYR=0.13198; LEU=0.00022; ASN=0.11292; PRO=0.00194; ARG=0.19835;
r=ARG; n=91; a=CG; | I=0.00000; O=0.00000; N=0.00000; P=0.15142; | TYR=0.03443; ASN=0.00616; ARG=0.11083;
r=ARG; n=91; a=NE; | I=0.00000; O=0.00000; N=0.00000; P=0.29265; | TYR=0.05905; ASN=0.07236; PRO=0.00043; ARG=0.16081;
r=ARG; n=91; a=CZ; | I=0.00000; O=0.00000; N=0.00000; P=0.37364; | TYR=0.08610; ASN=0.13015; PRO=0.00204; ARG=0.15535;
r=ARG; n=91; a=NH2; | I=0.00000; O=0.00000; N=0.00000; P=0.39548; | TYR=0.05916; ASN=0.21369; PRO=0.00470; ARG=0.11793;
r=ARG; n=91; a=CA; | I=0.00000; O=0.00000; N=0.00000; P=0.02902; | TYR=0.00694; ARG=0.02208;
r=ARG; n=91; a=N; | I=0.00000; O=0.00000; N=0.00000; P=0.01794; | TYR=0.00351; ARG=0.01443;
r=ARG; n=91; a=C; | I=0.00000; O=0.00000; N=0.00000; P=0.02592; | TYR=0.01928; ARG=0.00664;
r=ARG; n=91; a=O; | I=0.00000; O=0.00000; N=0.00000; P=0.05331; | TYR=0.04833; ARG=0.00498;
